New and innovative tools for accurate and in-depth aquaculture diagnostics, based on sequencing technology and the unique Next generation Sequencing platform (NGS) from PHARMAQ Analytiq.

An increasing degree of salmonid production (both smolt and downstreams poriuction) relies on closed or semi-closed production in water recirculation facilities.
Control of water quality is crucial to maintain stable production conditions. Little to no information regarding the microbiota in the water is known, but it is expected that it plays a vital role in affecting fish health, and welfare.
In cases of complex, and multi-factorial health challenges, where the direct cause is unknown or uncertain, current diagnostic tools often fall short.
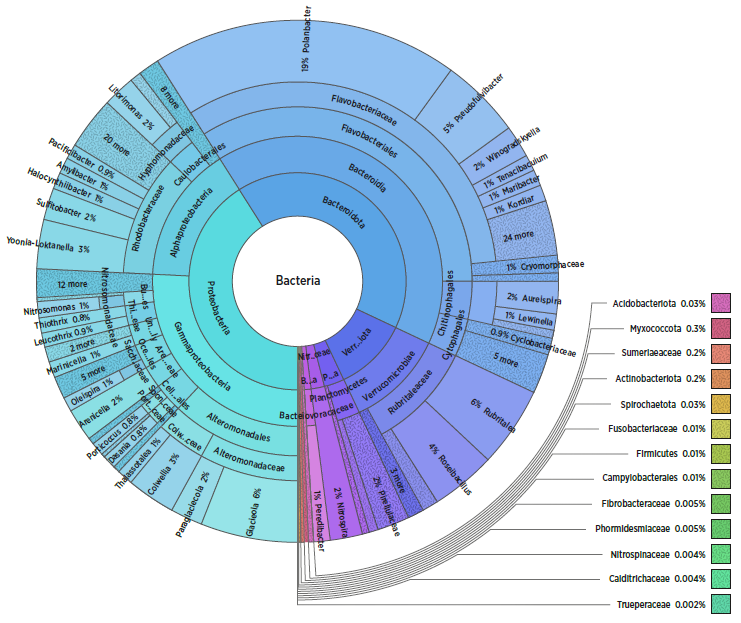
Figure 1. Sample data showing microbial profile from a water sample in a production facility.

An important, but under-utilised aspect of water quality is microbial composition, and how this affects, and contributes to fish health and welfare in production. The MONMIC project, led by SINTEF, has shown
that knowledge from one production facility is not necessarily directly applicable in other sites. This variation also makes it challenging to define what the normal flora at a specific site is or should be. To compensate for this, our goal has been to relate microbial profiles from water and biofilters to relevant parameters within water chemistry, site production data and health data from a wide range of production site types and forms.
Identify relative levels (%) of top 10 occurring bacterial species from intake water samples
Characterisation of microbial profi les in theintake water indirectly refl ects what you can expect from the bacterial composition in water in the tanks, and also indicates how effective particle removal and/or UV treatment in the site have been.
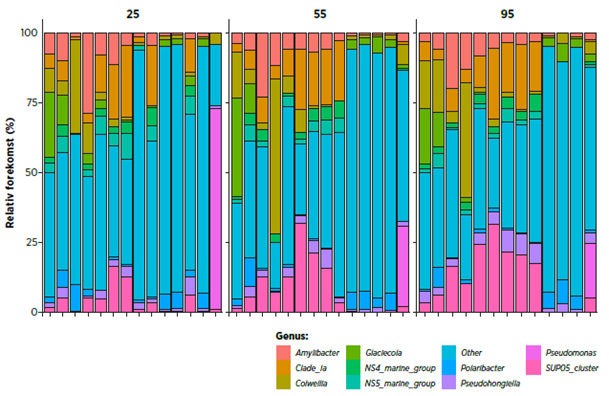
Figure 2. Sample data from intake water. Shows relative levels (%) of the 10 most frequently occurring bacterial species on Genus- level, for samples taken at different water depths
Identify relative levels (%) of different nitrifying bacteria from biofilter samples
Investigating the microbial status of biofilters gives an overview of all bacterial species present in the biofilter(s) AND enables identification of relative levels of nitrifying bacteria present. This can be used as an indicator of filter stability, normally represented by high bacterial diversity and low impact of external changes to this diversity.
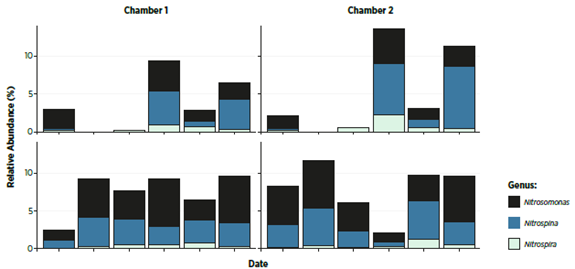
Figure 3. Sample data from biofi lter profi ling. Shows relative levels (%) of different nitrifying bacteria present on Genus- level, from biofilter samples.
Accurately identify bacteria using water samples taken from the tank
Identifying bacterial communities, and profiles, in water samples taken from the tanks themselves is also possible, in much the same
way as for intake water and biofilter. This also enables rapid identification of relative levels of potential pathogenic bacteria, as well as relative levels of potential geosmin producing bacteria present.
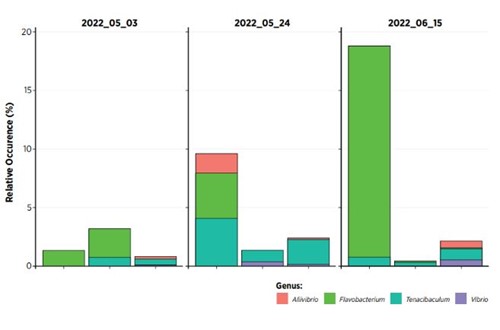
Figure 4. Sample data from water samples in a production site. Shows relative level (%) of bacteria known to contain species associated with disease in salmonids.
By comparing results from several different systems, with their unique variations in temperature, salinity, buffering systems, degree of oxygenation, disinfection a.s.o., you can start to build a broad and sound foundation, both for defining what the “normal flora” at a site should look like, and to identify which production parameters that mainly affect microbial composition in a positive and/or negative direction.
Challenges with skin ulcers andgilldisease are among the most important causes of loss in production in Norwegian aquaculture. In bothcases the disease picture is often complex withmultiple pathogens present and causal link to disease unresolved. In diagnostics, the focus has mainly been limited to documentation of pathogens present,
and to a very small degree on how the total microflora on the host potentially contributes to the diseases. Mapping and characterising total microbial flora on skin, skin ulcers and gills could be key to move forward and get a better overviewof which factors directly contribute to development of disease, and which only occur because of disease.
For both skin, and gill related flora and issues, we expect that a signifi cant amount of data, both in the form collection and analysis, is needed to document and identify correlations between microbial profi les and disease status.
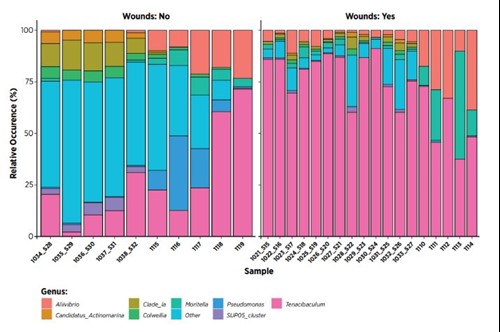
Figure 5. Sample data from swabs collected from fish with and without skin ulcers. Shows relative levels (%) of bacteria present.

The importance of applied use of data from full genome sequencing of viruses, and bacteria has become common knowledge after the Covid19 pandemic, where changes observed in the genome could have an effect of the success of the ongoing vaccine programs. For fish in aquaculture production, the situation and current challenges are in many ways quite similar, with pathogenic viruses, and bacteria constantly changing. Most salmonids in production are vaccinated against 5-7 agents of disease. Vaccine companies need to continuously monitor whether their vaccines cover the current circulating viruses, and bacteria, or not.
NGS based characterisation has the potential to give more detailed knowledge on which bacteria and/or viruses are present in Norwegian aquaculture on a much larger scale than before.
Figure 6. Sample data showing a graphic representation of a
bacterial genome from a fi eld sample of a bacterium
Combining genetic information from pathogens with geographical occurrence and production types, enables us to develop tools for molecular tracing of transmission routes and origin of infection. This knowledge can be crucial for introducing and developing correct preventive measures in production to combat disease.
PHARMAQ is the global leader in vaccines and innovation for aquaculture and part of Zoetis, the world leader in animal health. The company provides environmentally sound, safe and efficacious health products to the global aquaculture industry through targeted research and the commitment of dedicated people. Production facilities, administration and research and development activities are based in Norway with subsidiaries in Chile, United Kingdom, Vietnam, Turkey, Spain, Panama and Hong Kong. PHARMAQ has approximately 300 employees. The company's products are marketed in Europe, North and South America, and Asia. For further information, visit the company's website at www.pharmaq.com. Privacy Policy. Cookie Policy. Terms of Use.
Copyright © 2020 Zoetis LLC. All rights reserved.